As you recall from the configuration notes page, the deid recipe allows you to configure both cleaning of pixels and changing header values. This document covers the second - how to get, update, and replace header values.
Application Flow
Cleaning headers in dicom images typically has four parts:
- define a set of rules for updating values
- get current fields (if you need to use them to look up replacements, etc)
- update these fields as you see needed
- put (possibly updated) identifiers back into the data, and deidentify fully.
this document will talk about the first step in this process, how you can configure rules for the software. If you are interested in the command line client for these commands (and not functions) you should read about the client.
Defaults
The application does the following, by default, taking a conservative de-identification process:
- All fields are returned to you for inspection.
- You can replace none or all of these fields with your identifiers of choice
- The data will be rewritten with your changes, and all other fields will be blanked.
- A header field will be added that says the data has been anonymized.
However, you might want to do either of the following:
- have a specific action for some set of headers, where actions include
BLANK,REPLACE,JITTER,REMOVE,ADDandKEEP - perform some custom functions between
get,update, andput.
We will show you a working example of the above as you continue this walkthrough. For now, let’s review configuration settings.
Standard Anonymization
For a best effort anonymization, you will likely want to extract data, replace some fields, and put back the replacements. In this case you need to make a config file that should be provided with your function calls.
The configuration files for deid are called “deid recipes”
The format of this file is discussed below, and can be used to specify preferences for different kinds of datasets (dicom or nifti) and things to identify (pixels and headers).
Rules
You can create a specification of rules for the application to customize its behavior.
The file standard that I’ve been using is to call all files deid and use a reverse extension to indicate tag,
so deid.dicom is a deid recipe for dicom, and deid.chest-xray might be a recipe for chest xray.
deid.dicom.chest-xray could combine those two things, suggesting a recipe for dicom chest xrays.
Let’s look at an example:
FORMAT dicom
%header
ADD PatientIdentityRemoved YES
BLANK OrdValue
KEEP Modality
REPLACE id var:entity_id
JITTER StudyDate var:entity_timestamp
REMOVE ReferringPhysicianName
In the above example, we tell the application exactly how to deal with header fields for dicom.
We do that by way of sections (the lines that begin with % like %header and actions (eg, KEEP).
The fields above can include those that are specified in the file-meta section of the dicom, which are a unique namespace.
Each of these variables will be discussed in detail, next.
Format
The first thing that should appear in a recipe file is the FORMAT label. This is a message to the
application that the following commands are intended for dicom files, and the name dicom
matches exactly with the “dicom” module in the deid module.
Sections
Each section corresponds to a part of the data (eg, header or pixels) and then defines actions that can be taken for it.
Actions
Although different sections can have their own actions defined, for simplicity many sections share the same set:
- ADD
- BLANK
- JITTER
- KEEP
- REMOVE
- REPLACE
And the command in the file will either have the format of <ACTION> <FIELD> <VALUE> or
in the case of binary actions, just <ACTION> <VALUE>. For example, both of the following are valid:
#<ACTION> <FIELD> <VALUE>
ADD PatientIdentityRemoved YES
#<ACTION> <FIELD>
KEEP PixelData
Protected Fields
To protect you from replacing fields that might have un-intended consequences for the integrity of the dicom, by default we do not let you change a set of protected fields.
PixelDataRedPaletteColorLookupTableDataGreenPaletteColorLookupTableDataBluePaletteColorLookupTableDataVOILUTSequence
These fields are found within the header, and since we load the file metadata as well, we also protect the following:
FileMetaInformationGroupLengthFileMetaInformationVersionTransferSyntaxUIDImplementationClassUID
If you are using replace_identifiers, get_identifiers, or the DicomParser,
you can provide the boolean disable_skip=True to not skip any protected fields.
If you want to modify the set of skipped fields, then you can create your own
config.json and provide
the path as the config argument to either of these functions/classes.
Dynamic Values
var
In the case that your need to do something like “replace FIELD with my other variable,” then you want to format the value to tell the application that it should find the field in the data structure you pass it (discussed later). That format looks like this:
#<ACTION> <FIELD> <VALUE>
REPLACE PatientID var:suid
In the above, we tell the software to replace the field PatientID with whatever
value is defined under variable suid. Now let’s talk about how the actions are
relevant to different sections, first the header.
func
It might also be the case that you want to replace a field value with something generated on the fly from a function. In this case, you should define a “func:function_name” instead:
#<ACTION> <FIELD> <VALUE>
REPLACE PatientID func:generate_suid
When you do this, the function is expected to be defined in the customized
item dictionary that you pass in (e.g., modified output from get_identifiers)
See the Frame of Reference example
for a walkthrough of how to do this. Note that you can equivalently do this with
ADD to add a value based on a function:
#<ACTION> <FIELD> <VALUE>
ADD MyNewField func:generate_field
For both of the above, the function should return a value that you want replaced
or added. In the case of wanting to remove something based on conditional logic
that you define in a function, you can use a function with REMOVE:
#<ACTION> <FIELD> <VALUE>
REMOVE ALL func:is_name
The function should return a boolean (True or False) to indicate if you want the field in question removed. For the above, all fields would be checked against your function to determine removal status. Here is an example function that grabs a patient name, and then checks fields to see if the name is present. Fields where it is present will be removed (return True).
def is_name(dicom, value, field):
name = dicom.get('PatientName')
currentvalue = dicom.get(field)
splitvalues = name.split('^')
for phi in splitvalues:
if len(phi) > 4 and phi in currentvalue:
return True
return False
Note that “value” refers to the value from the recipe, in this case, the full func:is_name.
Thus, we retrieve the current value by getting the field from the dicom, and then
check if any portion of the name is represented in the current value. We return
True if this is the case. The dicom is the dicom file (read in with Pydicom) that you can use to interact
with (in the example above we grab the PatientName).
Header
We know that we are dealing with functions relevant to the header of the image
by way of the %header section. This section can have a series of commands called
actions that tell the software how to deal with different fields. For the header
section, the following actions are allowed, and each is specific to an action to
be taken on a header field/value:
- ADD: Add a new field to the dataset(s). If the value is a string, it’s assumed to be the value that is desired to be added. If the value is in the form
var:OrdValuethen the application will expect to find the value to replace in a variable in the request calledOrdValue(more on this later). - BLANK: If you want to blank a field instead of remove it, use this option. This is the default action.
- KEEP: implies that the value should not be replaced, jittered, removed, or blanked.
- REPLACE: implies that the value should be replaced by a string, or a variable in the format
var:FieldName. - JITTER: implies that the value should be jittered (“deviated”) by a string, or a variable in the format
var:FieldName. - REMOVE: completely remove the field from the dataset (if not protected by
KEEP, changed by ‘REPLACE’ or ‘JITTER’, or is in default skip list).
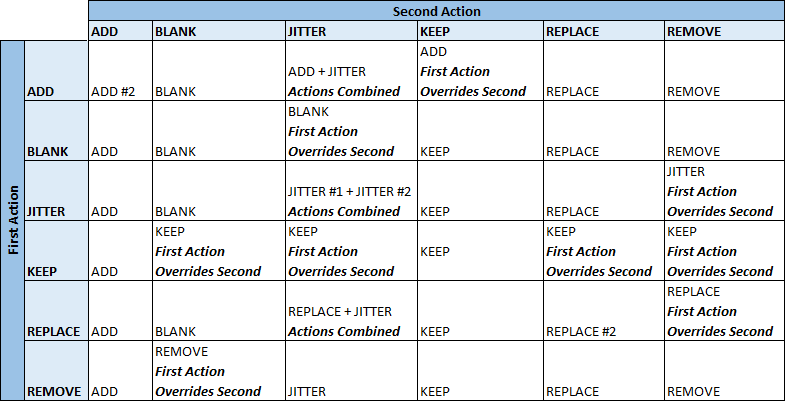
For the above, given that there are conflicting commands, the more conservative is given preference. For example:
KEEP > ADD > REPLACE > JITTER > REMOVE > BLANK
For example, if I add or keep a header, and then also specify to blank or remove it, it will be kept. If I specify to blank a header and remove it, it will be removed. If I specify to replace a header and blank it, it will be blanked. Most of the time, you won’t need to specify remove, because it is the default. If we were to come up with a pretend config file to represent the default, it would look like this:
FORMAT dicom
%header
ADD PatientIdentityRemoved YES
REMOVE ALL
KEEP PixelData
KEEP SamplesPerPixel
KEEP Columns
KEEP Rows
The above would remove everything except for the pixel data, and a few fields that are relevant to its dimensions. It would add a field to indicate the patient’s identity was removed.
The table below shows the full details of how multiple actions defined on the same field will interact with each other. In general the second action encountered on a field will supercede the first, however there are a few cases in which the actions are combined or the first supercedes the second.
Jitter
For jitter, you can add a hard coded number, or a variable to specify it:
JITTER StudyDate var:jitter
JITTER Date 31
JITTER PatientBirthDate -31
Field Expansion
In some cases, it might be extremely tenuous to list every field ending in the same thing, to perform the same action for. For example:
JITTER StudyDate var:jitter
JITTER Date var:jitter
JITTER PatientBirthDate var:jitter
could much better be captured as:
JITTER endswith:Date var:jitter
and this is the idea of an expander. And expander is an optional filter
applied to a header field (the middle value) to select some subset of header
values. Currently, we support startswith, endswith, contains, select,
allexcept, and allfields.
The following examples show what fields are selected based on each filter. For all examples, the test is done making the values lowercase.
endswith
The endswith filter will look for header field names that end with a particular expression.
Lower and upper casing does not matter, so writing endswith:Date is akin to writing
endswith:Date.
JITTER endswith:Date var:jitter
['AcquisitionDate', 'ContentDate', 'InstanceCreationDate', 'PatientBirthDate', 'PerformedProcedureStepStartDate', 'SeriesDate', 'StudyDate']
startswith
The startswith filter will look for header field names that start with a particular expression. Casing also doesn’t matter.
REMOVE startswith:Patient
['PatientAddress', 'PatientAge', 'PatientBirthDate', 'PatientID', 'PatientName', 'PatientPosition', 'PatientSex']
contains
The contains filter searches for a string of interest in the field. For example, if we ask for fields that contain “Name” will look for header field names that contain the string “Name” in any casing.
BLANK contains:Name
['InstitutionName', 'NameOfPhysiciansReadingStudy', 'OperatorsName', 'PatientName', 'ReferringPhysicianName']
Notice how we get Name in uppercase (when our search string was lowercase) and it can appear anywhere in the field.
select
The select filter can be used to select fields based on properties
of the DICOM element. These filters have the form
select:property:value.
Currently implemented property filters are select:group: to select
elements by tag group and select:VR: to select elements by value
representation.
The select:group: filter selects fields based on their DICOM
group. Groups values for the filter are specified as hexadecimal
numbers (of up to four digits). For example, the following rules will
keep all elements contained within DICOM groups 0x0018, 0x0020 and
0x0020:
KEEP select:group:0018
KEEP select:group:0020
KEEP select:group:0028
The select:VR: filter selects fields based on the value
representation (VR) of the element. VR values are specified as the two
character strings. For example, the following rules will blank all
elements that have a VR of TM (time):
BLANK select:VR:TM
all
All fields will allow you to apply a filter to all fields. In the example below, we have a function “remove_identifiers” defined in the python environment, and will run it over all fields, returning a value to update the field in question.
ADD ALL func:remove_identifiers
except
Akin to all fields, except provide a pattern that you want to exclude from a global selector. Here are some examples to include all fields except StudyDate or StudyTime.
BLANK except:StudyDate|StudyTime
BLANK except:StudyTime
except is checking field names against given pattern, so if you for example set
REMOVE except:Manufacturer
following fields will be preserved: Manufacturer and ManufacturerModelName
If you are familiar with regular expressions, you’ll notice the “|” which means “or” in the regular expression. You are free to write whatever regular expression fits your needs to disclude particular fields here.
Does this include private tags?
If you choose to not remove private tags, as of version 0.0.41 of deid, you can also search private tag identifiers based on a number. Here is an example of private tags defined for a dicom file:
[(0011, 0003) Private Creator AE: 'Agfa DR',
(0019, 0010) Private Creator LO: 'Agfa ADC NX',
(0019, 1007) Private tag data CS: 'YES',
(0019, 1021) Private tag data FL: 6.039999961853027,
(0019, 1028) Private tag data CS: 'NO',
(0019, 1030) Private tag data LT: '',
(0019, 10f5) [Cassette Orientation] CS: 'LANDSCAPE',
(0019, 10fa) Private tag data IS: "297",
(0019, 10fb) Private tag data FL: 2.4000000953674316,
(0019, 10fc) Private tag data IS: "171",
(0019, 10fd) Private tag data CS: 'NO',
(0019, 10fe) [Unknown] CS: 'MED']
If we wanted to selected those that included 0019, we could do the following:
REMOVE contains:0019
The underlying function that expands the expression would return a subset of these tags - these are just the tag keys that are used to index the dicom structure:
from deid.dicom.fields import expand_field_expression
from deid.dicom.tags import get_private
# contenders should include all fields plus private tag names as keys
contenders = dicom.dir() + [e.tag for e in get_private(dicom)]
fields = expand_field_expression(dicom=dicom, field="contains:0019", contenders=contenders)
The result would be the following:
[(0019, 0010),
(0019, 1007),
(0019, 1021),
(0019, 1028),
(0019, 1030),
(0019, 10f5),
(0019, 10fa),
(0019, 10fb),
(0019, 10fc),
(0019, 10fd),
(0019, 10fe)]
Which then can be used as indices to the dicom to get the full private tag.
[dicom.get(tag) for tag in fields]
[(0019, 0010) Private Creator LO: 'Agfa ADC NX',
(0019, 1007) Private tag data CS: 'YES',
(0019, 1021) Private tag data FL: 6.039999961853027,
(0019, 1028) Private tag data CS: 'NO',
(0019, 1030) Private tag data LT: '',
(0019, 10f5) [Cassette Orientation] CS: 'LANDSCAPE',
(0019, 10fa) Private tag data IS: "297",
(0019, 10fb) Private tag data FL: 2.4000000953674316,
(0019, 10fc) Private tag data IS: "171",
(0019, 10fd) Private tag data CS: 'NO',
(0019, 10fe) [Unknown] CS: 'MED']
These notes are provided to give detail about the implementation - you do not need to worry about using these underlying functions to do expansion, only that they are working to expose even private tags for parsing.
Example
The suggested approach that you should take, replacing the main entity data with some identifier that you’ve selected, would look something like this:
FORMAT dicom
%header
ADD PatientIdentityRemoved YES
REPLACE PatientID var:id
If you wanted to also replace the image (SOPInstanceUID) with an identifier, that might look like this:
FORMAT dicom
%header
ADD PatientIdentityRemoved YES
REPLACE PatientID var:id
REPLACE SOPInstanceUID var:source_id
And the expectation would be that you provide variables with keys source_id and id
appended to the response from get that is handed to the put action.
Future Additions
Format nifti
In the future when we add support for other data types, the config might look something like this (note the added nifti section):
FORMAT dicom
%header
ADD PatientIdentityRemoved YES
REPLACE PatientID var:id
REPLACE InstanceSOPUID var:source_id
FORMAT nifti
%header
ADD PatientIdentityRemoved YES
REPLACE PatientID var:id
REPLACE InstanceSOPUID var:source_id
Value Expansion
These same filters can also be used with any action that is considered a boolean,
for example, the REMOVE tag. As we showed previously, you can remove using
a filter like “contains” to select some subset of fields:
REMOVE contains:Patient
which would remove all fields that contain “Patient.” What if we want to perform this same kind of check, but with a value? For example, let’s say that we have a regular expression to describe a number, and we want to remove any field that matches. We could do:
REMOVE ALL contains:(\d{7,0})
Would parse through ALL fields, and remove those that contain a match to the regular expression. All supported expanders include:
- contains
- notcontains
- equals
- notequals
Now that you know how configuration works, you have a few options. You can learn how to define groups of tags based on fields or values in groups, or if you want to write a text file and get going with cleaning your files, you should look at some examples for generating a basic get. This is the action to get a set of fields and values from your dicom files. For a full walk through example with a recipe, see the recipe example
